Bradley Davidson ’91

Research | People | Publications
Courses
BIOL 1: Cellular & Molecular Biology
Team taught. An introduction to the study of living systems illustrated by examples drawn from cell biology, biochemistry, genetics, microbiology, neurobiology, and developmental biology.
BIOL 24: Developmental Biology
In this course, we will explore the process by which single cells (fertilized eggs) develop into complex organisms. Students will conduct detailed observations of live embryos and engage in independent experimental analysis during weekly laboratory sessions.
BIOL 125: Frontiers in Developmental Biology
Through discussion of the primary literature and independent experimental studies, students will investigate current gaps in our understanding of animal development. Potential topics include: the interplay between embryonic development and evolution; how gene regulatory networks generate complex patterns of cell identity; and the ability of cells to interpret their environment using dynamic internal structures.
BIOL 128: Evolution and Development
In this course, we will explore how alterations in embryonic development contribute to evolution. We will cover a wide range of examples spanning body plan diversification during the Cambrian explosion through the much more recent diversification of humans and other primates. Through engagement with the primary literature, students will learn how comparative genomics, experimental analysis of gene regulatory networks and in-depth dissection of cellular processes have revealed discrete genetic, molecular and cellular changes underlying the evolutionary acquisition of novel traits.
Research
Signaling between embryonic cells and heart formation
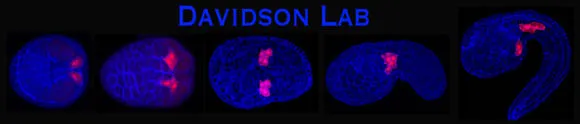
The heart founder lineage labeled with RFP (red fluorescent protein) in a series of staged Ciona embryos
About 40,000 children are born with heart defects every year. In order to treat these conditions we must gain a comprehensive understanding of heart formation, including the very first steps taken by heart cells in early embryos. We study this question in the model organism Ciona intestinalis. Ciona embryos are very simple both cellularly and genetically and yet they are closely related to the vertebrates, using many of the same heart genes as human embryos. This simplicity allows us to observe the exact behavior of individual heart cells and study how heart genes influence these behaviors. Our goal is to use what we learn in Ciona to guide heart research in other model organisms such as frogs, chicken and mice, eventually providing the framework required for the diagnosis and treatment human heart defects.
Research Interests:
In a vertebrate embryo, cells are first instructed to form heart by a complex array of signals from neighboring cells, including Fibroblast Growth Factors (FGFs). Complex interactions between these signals have made it difficult to tease out the specific contribution of individual factors. This problem is exacerbated by redundancy in these pathways. For example, there are four vertebrate FGF receptors with numerous isoforms and approximately 20 distinct FGF ligands. Furthermore, each of these factors coordinates multiple aspects of development; influencing cell behavior as well as regulatory gene expression. Deciphering how signaling factors direct discrete cell behaviors is challenging in the dense architecture of vertebrate embryos. In order to side-step some of these challenges we have initiated studies of heart development in the sea squirt, Ciona intestinalis.
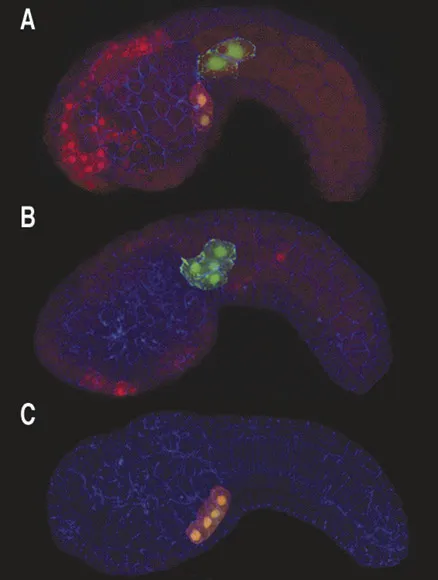
In these embryos potential heart cells are labeled by green fluorescent protein, FGF signaling causes two of these cells to form heart (and express a red fluorescent reporter gene)
Ciona is a chordate and thus a member of our own phyla. This close evolutionary relationship makes Ciona genetically more similar to humans than other invertebrate model organisms. The single-chambered Ciona heart forms by similar processes as the vertebrate heart tube, but in the context of a highly simplified embryo (see banner image above). Ciona has only one copy of many key developmental genes that were later duplicated in the vertebrates, allowing stringent assessment of gene function. Additionally, low cell numbers permit unprecedented visualization of cell morphology and migration. Despite this simplicity, the fundamental program for early vertebrate heart development is conserved in Ciona.
Currently, our research is focused on revealing the precise function of FGF in early heart development. We have demonstrated that FGF signaling causes a group of 4 founder cells to undergo an asymmetric division. The smaller daughters of this division respond to continued FGF signaling by activating heart genes and migrating towards the site of future heart formation while the larger daughter form tail muscle.
Through transgenic manipulations, we can disrupt FGF signaling specifically in these four cells, blocking heart development. Conversely, we can activate downstream factors and cause the entire group of cells to migrate and form extra heart tissue (above image). We are also able to isolate Ciona heart cells and examine lineage-specific gene expression. This analysis employs micro-arrays designed to probe all predicted coding regions in the Ciona genome. Through these techniques we have identified an extensive set of heart genes up-regulated by FGF. Future studies will focus on determining the role of these FGF target genes in heart development as well as identifying the precise transcriptional mechanisms by which FGF and downstream factors co-ordinate heart gene expression.
We are also interested in understanding the differential response of founder cells to FGF signaling. Why do the smaller daughter cells respond while the larger daughters do not? Our latest results indicate that this differential response is dictated by a remarkable invasion of neighboring epidermal (skin) cells. We are currently investigating how this invasive behavior boosts FGF signaling and heart gene activation. This newly characterized 'invasive signaling' mechanism has intriguing implications for vertebrate heart development and may also be involved in cancer cell metastasis.
People In The Lab

Sydney Popsuj Postdoctoral Research Associate |
Publications
Cota, C. D., ^Dreier, M., ^Colgan, W., ^Cha, A., ^Sia, T., & Davidson, B. Cyclin-dependent Kinase 1 and Aurora Kinase choreograph mitotic storage and redistribution of a growth factor receptor Plos Biology. 2021 .\ PMCID: PMC7808676; PMID: 3395410
Westerman, E. L., Bowman S, Davidson, B, Davis, M, Larson, E and Stanford, C. (2020) Deploying Big Data to Crack the Genotype to Phenotype Code. Integrative and Comparative Biology. doi:10.1093/icb/icaa055
DeBiasse, M. B., ^Colgan, W. N., ^Harris, L., Davidson, B. & Ryan, J. F. (2020) Inferring Tunicate Relationships and the Evolution of the Tunicate Hox Cluster with the Genome of Corella inflata. Genome Biology and Evolution 12, 948–964.
Dardaillon, Justine; Dauga, Delphine; Simion, Paul; Faure, Emmanuel; Onuma, Takeshi; DeBiasse, Melissa; Louis, Alexandra; Nitta, Kazuhiro; Naville, Magali; Besnardeau, Lydia; Reeves, Wendy; Wang, Kai'.; Fagotto, Marie; Guéroult-Bellone, Marion; Fujiwara, Shigeki; Dumollard, Rémi; Veeman, Michael; Volff, Jean-Nicolas; Roest Crollius, Hugues; Douzery, Emmanuel; Ryan, Joseph; Davidson, Bradley; Nishida, Hiroki; Dantec, Christelle; Lemaire, Patrick (2019). ANISEED 2019: 4D exploration of genetic data for an extended range of tunicates. Nucleic Acids Research 439, 965–8.
^Colgan, W., ^Leanza, A., ^Hwang, A., DeBiasse, M., ^Rodrigues, D., ^Llosa, I., ^Adhikari H., ^Barreto Corona G., ^Bock S., ^Carillo-Perez A., ^Currie M., ^Darkoa-Larbi S., ^Dellal D., ^Gutow H., ^Hokama P., ^Kibby E., ^Linhart N., ^Moody S., ^Naganuma A., ^Nguyen D., ^Stanton R., ^Stark S., ^Tumey C., ^Velleca A., Ryan, J., Davidson B. (2019) Variable levels of drift in tunicate cardiopharyngeal gene regulatory elements. EvoDevo 1–17 (2019). doi:10.1186/s13227-019-0137-2
^Palmquist, K., & Davidson, B. (2017). Establishment of lateral organ asymmetries in the invertebrate chordate, Ciona intestinalis. EvoDevo 8(12), 1-16. DOI 10.1186/s13227-017-0075-9
Cota, C. D., ^Palmquist, K. & Davidson, B. (2017). Heart development in Ciona. Reference Module in Life Sciences
Segade, F., Cota, C. D., ^Famiglietti, A., ^Cha, A. & Davidson, B. (2016). Fibronectin contributes to notochord intercalation in the invertebrate chordate, Ciona intestinalis. EvoDevo 7(21), 1–16. doi:10.1186/s13227-016-0056-4
Cota, C. D., & Davidson, B. (2015). Mitotic membrane turnover coordinates differential induction of the heart progenitor lineage. Developmental Cell, 34(5), 505–519. doi: 10.1016/j.devcel.2015.07.001
Cota, C. D., Segade, F., & Davidson, B. (2013). Heart genetics in a small package, exploiting the condensed genome of Ciona intestinalis. Briefings in functional genomics. doi:10.1093/bfgp/elt034
Norton, J. Cooley, J. Islam, T., Cota, C. and Davidson, B. (2013) Matrix adhesion directs asymmetric heart progenitor specification in the basal chordate, Ciona intestinalis. Development, 140(6), 1301-11. [DOI:10.1242/dev.085548]
Woznica, A., Haeussler, M., Starobinska, E., Jemmett, J., Li, Y., Mount, D., & Davidson, B. (2012). Initial deployment of the cardiogenic gene regulatory network in the basal chordate, Ciona intestinalis. Developmental Biology, 368(1), 127–139. [DOI:10.1016/j.ydbio.2012.05.002]
Cooley, J., Whitaker, S., Sweeney, S., Davidson, B. (2011) Cytoskeletal polarity mediates localized induction of the heart precursor lineage. Nature Cell Biology. 13, 952-7 [DOI: 10.1038/ncb2291]
Ragkousi, K., Beh, J., Sweeney, S., Starobinska, E., Davidson, B. (2011) A single GATA factor plays discrete, lineage specific roles in ascidian heart development. Developmental Biology 352,154-63
Sobreira, T. J., Marletaz, F., Simoes-Costa, M., Schechtman, D., Pereira, A. C., Brunet, F., Sweeney, S., Pani, A., Aronowicz, J., Lowe, C. J. et al. (2011) Structural shifts of aldehyde dehydrogenase enzymes were instrumental for the early evolution of retinoid-dependent axial patterning in metazoans. Proc Natl Acad Sci U S A 108, 226-31
José Xavier-Neto, Brad Davidson, Marcos Sawada Simoes-Costa, Rodrigo Abe Castro, Hozana Andrade Castillo, Allysson Coelho Sampaio and Ana Paula Azambuja (2010) Evolutionary Origins of Hearts in Heart Development and Regeneration. Editors; Rosenthal and Harvey. 2010
Christiaen, L., Stolfi A., Davidson B. and Levine. M. (2009) Spatio-temporal intersection of Lhx3 and Tbx6 defines the cardiac field through synergistic activation of Mesp. Dev Biol 328(2), 552-60
Christiaen L., Davidson B., Kawashima T., Powell W., Nolla H., Vranizan K., Levine M. (2008) The transcription/migration interface in heart precursors of Ciona intestinalis. Science 320,1349-1352. [DOI: 10.1126/science.1158170]
Davidson B. (2008) Movers and shakers: evolution and development of the mesoderm. J Exp Zoolog B Mol Dev Evol. 310:1-4.
Beh, J., Shi, W., Levine, M., Davidson B., Christiaen, L. (2007) FoxF is essential for FGF-induced migration of heart progenitor cells in the ascidian Ciona intestinalis. Development. 134: 3297-305.
Davidson, B. (2007) Ciona intestinalis as a model for cardiac development. Sem Cell Dev. 18: 16-26
Davidson, B., Shi, W., Beh, J., Christiaen, L. and Levine, M. (2006). FGF signaling delineates the cardiac progenitor field in the simple chordate, Ciona intestinalis. Genes Dev 20, 2728-38.
Davidson, B. and Christiaen, L. (2006). Linking chordate gene networks to cellular behavior in ascidians. Cell 124, 247-50.
Simoes-Costa, M. S., Vasconcelos, M., Sampaio, A. C., Cravo, R. M., Linhares, V. L., Hochgreb, T., Yan, C. Y., Davidson, B. and Xavier-Neto, J. (2005). The evolutionary origin of cardiac chambers. Dev Biol 277, 1-15.
Shi, W., Levine, M. and Davidson, B. (2005). Unraveling genomic regulatory networks in the simple chordate, Ciona intestinalis. Genome Res 15, 1668-74.
Davidson, B., Shi, W. and Levine, M. (2005). Uncoupling heart cell specification and migration in the simple chordate Ciona intestinalis. Development 132, 4811-8.
Johnson, D. S., Davidson, B., Brown, C. D., Smith, W. C. and Sidow, A. (2004). Noncoding regulatory sequences of Ciona exhibit strong correspondence between evolutionary constraint and functional importance. Genome Res 14, 2448-56
Davidson, B., Smith Wallace, S. E., Howsmon, R. A. and Swalla, B. J. (2003). A morphological and genetic characterization of metamorphosis in the ascidian Boltenia villosa. Dev Genes Evol 213, 601-11.
Davidson, B., Jacobs, M., and Swalla, B.J. (2003) The individual as a module: Metazoan evolution and coloniality. In: Modularity in Development and Evolution. Gerhard Schlosser and Günter Wagner, eds. University of Chicago Press.
Davidson, B. and Levine, M. (2003). Evolutionary origins of the vertebrate heart: Specification of the cardiac lineage in Ciona intestinalis. Proc Natl Acad Sci U S A 100, 11469-73.
Davidson, B. and Swalla, B. J. (2002). A molecular analysis of ascidian metamorphosis reveals activation of an innate immune response. Development 129, 4739-51.
Davidson, B. and Swalla, B. J. (2001). Isolation of genes involved in ascidian metamorphosis: epidermal growth factor signaling and metamorphic competence. Dev Genes Evol 211, 190-4.
Satoh, N., Satou, Y., Davidson, B. and Levine, M. (2003). Ciona intestinalis: an emerging model for whole-genome analyses. Trends Genet 19, 376-81.
back to Swarthmore Biology